1Extended CV Cycling
During the course of electrochemical research, a user may occasionally need to perform many cyclic voltammetry (CV)
experiments in a row. There are several applications where this kind of procedure may be required (e.g., electrochemical cleaning of a electrode, durability testing, or supercapacitor charge/discharge tests), and the exact procedure desired can vary greatly depending on the individual's research. For example, one researcher may simply need to conduct 500 CVs in a row to electrochemically clean an electrode surface, and they may not be interested in saving or examining any part of the data during or after the experiment. Another researcher, conversely, may wish to perform 500 CVs in a row and then need to analyze a voltammogram every so often (e.g., every 50th or 100th CV) to compare selected curves at the conclusion of the test.
This post details a few different strategies using the Pine Research AfterMath software
for performing long term CV experiments.
1.1Setting up CV Specification
For simplicity, a certain set of CV parameters has been assumed and applied to the strategies discussed herein. Substitution or alteration of any or all of these parameters should be done to meet the researcher's specific experimental design; however, the basic strategies can still be applied. The list of parameters used for demonstration in this post are shown below (see Table 1):
| Parameter |
Value |
| Experiment Type |
Cyclic Voltammetry (CV) |
| Number of CV Cycles |
1000 |
| Starting/Lower Potential |
-500 mV |
| Upper Potential |
800 mV |
| Sweep Rate |
100 mV/s |
Table 1. List of Parameters Used in this Post for Long Term CV Testing
The first step is to create a new archive in AfterMath
(see Figure 1). Once the archive has been created, a CV specification can be selected by either right-clicking on the archive, and selecting "New", then "Experiment", then "Cyclic Voltammetry (CV)". Alternatively, the specification can be clicked directly from the "Experiments" menu at the top of the screen (see Figure 2).
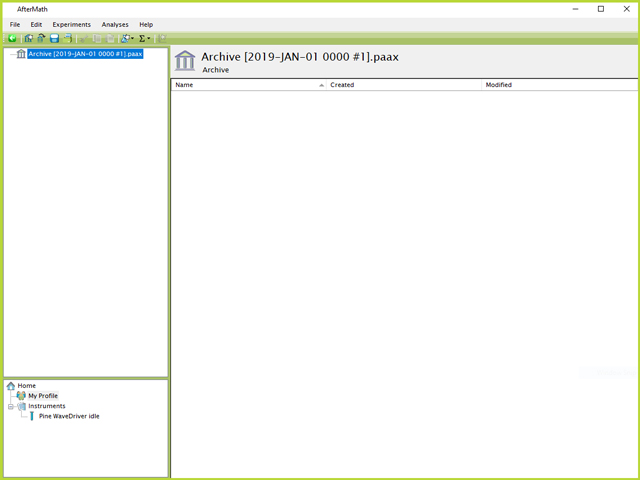
Figure 1. New Archive in AfterMath
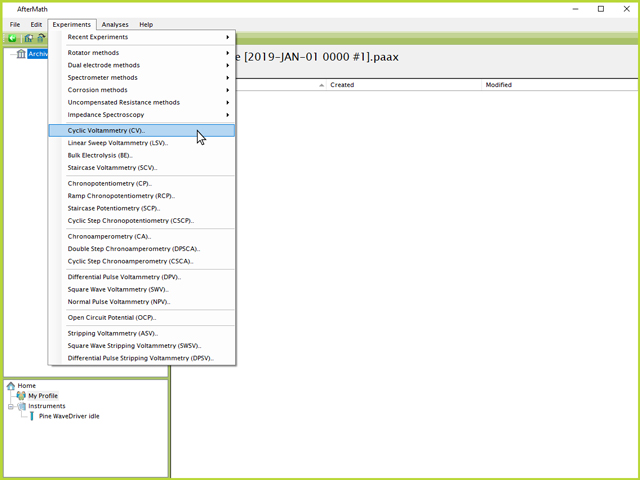
Figure 2. Selecting Cyclic Voltammetry (CV) from Top Experiments Menu in AfterMath
1.1.1Method 1: Single CV Specification
The first method that can be attempted for carrying out 1000 CVs in a row is to use a single CV specification and increase the number of segments. In this case, by starting and ending at the same lower limit (-500 mV, see Table 1), it means the number of segments needed is simply twice the number of CV cycles (2000, see Figure 3).
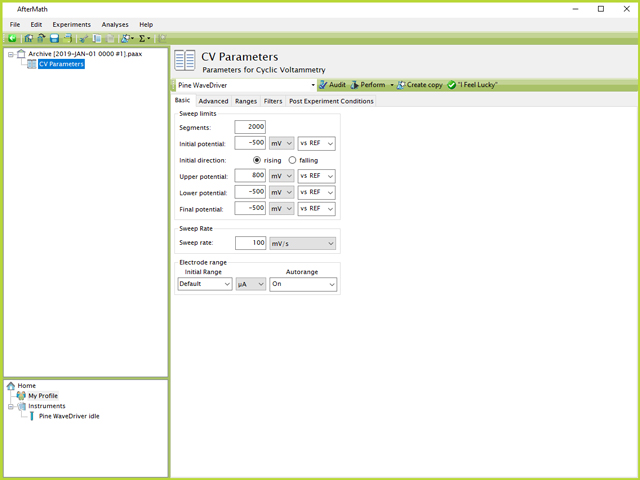
Figure 3. Single CV Parameters for 1000 Cycle Test in AfterMath
However, this approach may result in an error when attempting to Perform the experiment due to the large number of data points to be collected in a single experiment. AfterMath has a pre-defined limit that allows 10 million points per experiment. Therefore, if the combination of segments, limits, and threshold to record points will exceed this limit, an error message may occur (see Figure 4).
NOTE: When attempting to run a long number of cycles in a single experiment, an error message may appear that reads "Phase delay conflict".
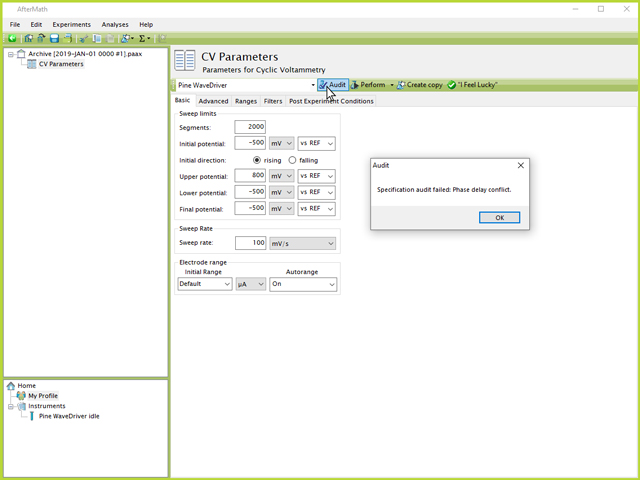
Figure 4. Single CV Audit Error from Too Many Points in AfterMath
By decreasing the number of segments by a factor of ten to 200, an audit of the experimental parameters shows that it can be successfully performed (see Figure 5):
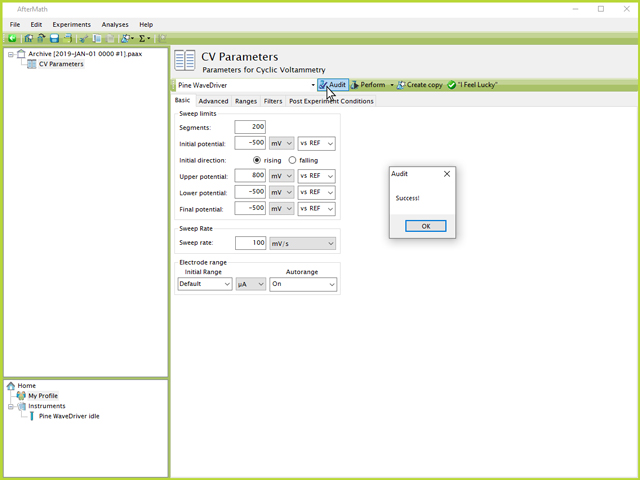
Figure 5. Successful CV Parameters Audit in AfterMath
To run the desired number of total CVs, the procedure shown in Figure 5 needs to be performed ten times. This can either be done by successively clicking the "Perform" button ten times (each subsequent action queues up a new experiment rather than overwriting or eliminating the previous/current one), or by using the small arrow next to the word "Perform" and directly choosing the "10x" option (see Figure 6). This is a more convenient option than clicking ten separate times because AfterMath automatically toggles to the active experiment plot following a click of the "Perform" button, meaning the user would need to repeatedly navigate back to the CV Parameters node.
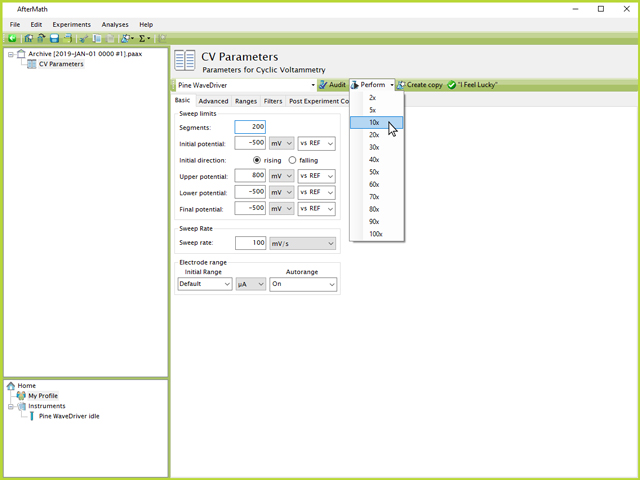
Figure 6. Perform 10x Option in AfterMath
1.1.2Method 2: Multiple CV Specifications using One Sequence
An alternative to using the same CV specification many times in a row is to use a Sequence.
From the steps shown in Section 1.1.1, it is clear that 200 segments (100 total CVs) is an appropriate and maximum size of individual CV experiments to conduct per specification node. Therefore, adding 10 individual CV specifications underneath a Sequence, each with the set of parameters shown in Figure 5, will result in 1000 CVs once the Sequence is performed (see Figure 7).
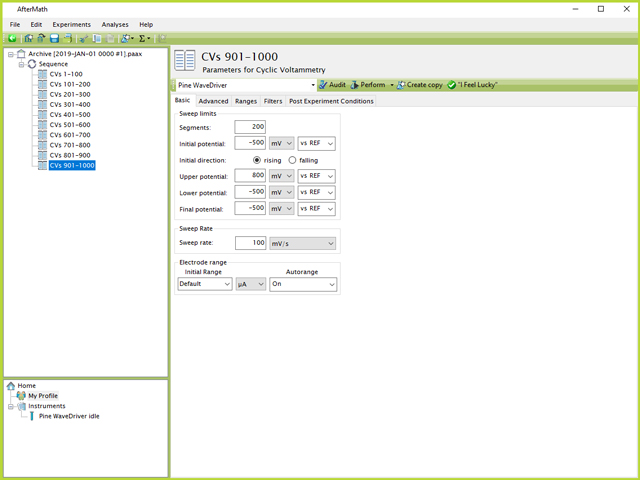
Figure 7. Sequence of CVs in AfterMath
An added benefit to using a Sequence is that each CV specification can be named according to the number of CVs that will be performed (e.g., CVs 401-500, see Figure 7). Much like an experiment specification, the entire sequence of experiments can be performed by clicking the Sequence node in the left tree structure, then clicking the "Perform Sequence" button on the upper-right side of the screen (see Figure 8). Additionally, on this screen the list and order of experiments to be executed in the Sequence is listed. This list may be drag and dropped to any desired order by the user.
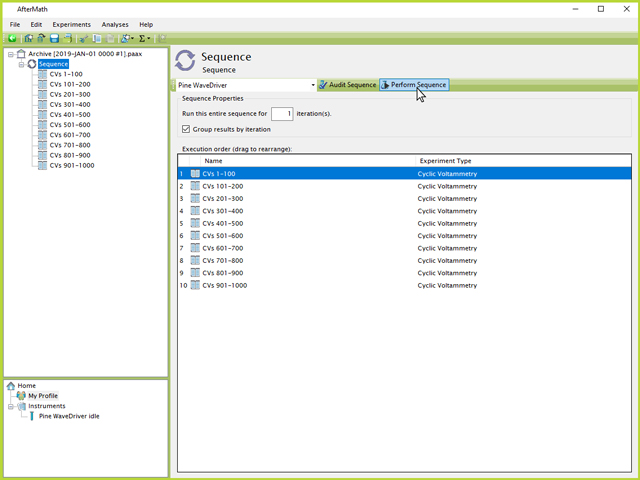
Figure 8. Perform Button on Sequence of CVs in AfterMath
1.1.3Method 3: Multiple CV Specifications using Multiple Sequences
One of the disadvantages of Methods 1 and 2 shown in Sections 1.1.1 and 1.1.2, respectively, is that all 1000 CVs are being performed from the same primary archive file. This means that during the entire duration of the experiment, a large volume of random access memory (RAM) must be allocated to AfterMath. Certain computers may have difficulty running programs that use up too high a percentage of the available RAM, which can occasionally result in a crash.
AfterMath does have built-in data recovery safeguards in the event there is a loss in power, or the computer/program crashes. However, it is generally best to not need to rely on a recovery since there may be incomplete data collected and the electrodes may also be damaged if an abrupt change in electrochemical state is induced by the instrument during such a failure.
Therefore, perhaps a more desirable technique for longer term experiments is to split up the number of cycles into multiple different Sequences across multiple different archives (see Figure 9). In this case, five separate archives are shown, each containing a Sequence with two CV specifications totaling 200 CVs. Experiments can be performed in AfterMath, in any order, independently of the archive in which they exist. Therefore, by successively clicking to each Sequence and clicking each "Perform Sequence" button, the 1000 CVs can be queued to the instrument.
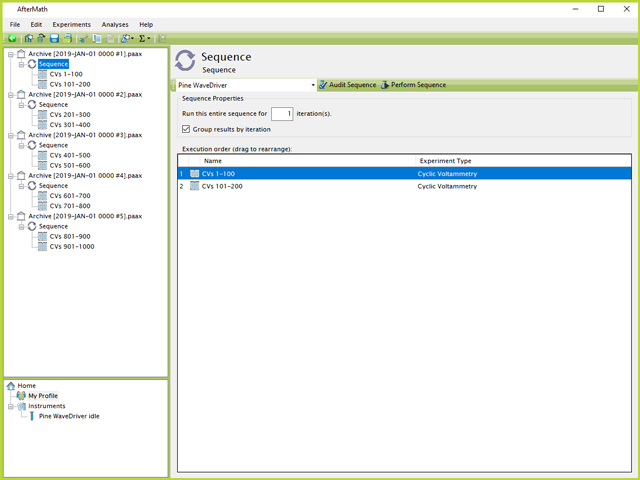
Figure 9. Five Archive File Split Sequences in AfterMath
The other added benefit of splitting the five Sequences into five separate archive files, as opposed to five Sequences inside the same archive file, is the ability to "Auto-close Archive". This option can be found by right-clicking on an archive node in the left tree structure and clicking the "Auto-close Archive" option (if the checkbox already appears, the option is already active and does not need to be clicked again, see Figure 10). This option will cause AfterMath to automatically save and close an archive once all experiments have completed, thus freeing up RAM space on the computer. In the example shown in Figure 10, once the first 200 CVs have finished, the first archive file will be saved to disk and closed to free up RAM so that the remaining experiments may run more smoothly and reduce risk of a crash.
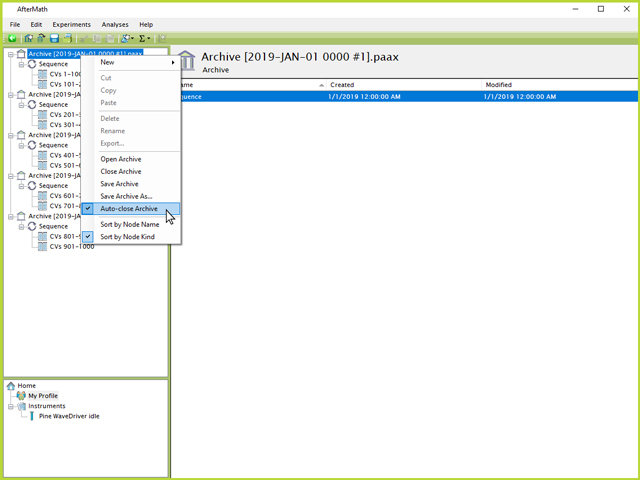
Figure 10. Auto-Close Archive Setting in AfterMath for Long Term Experiments
The above examples will all result in a total of ten CV results nodes, each containing 100 CVs. However, in the case a user needs to analyze the CV data every 100th cycle, for example, this method may make that task somewhat cumbersome. If the researcher knows beforehand which CVs are of greater interest, the CV specifications in each Sequence across all archives can be adjusted as shown in Figure 11:
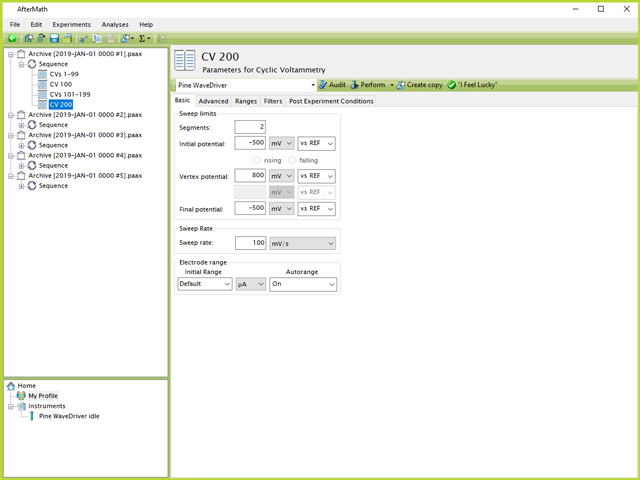
Figure 11. Sequence with Every 100th CV an Individual Experiment
In this example, the first 99 CVs will be collected in their own experiment, followed by a single cycle CV for number 100. When the user examines the data after it has completed, it will then be very easy to find and analyze the 100th CV because it will be properly named and independent from the other CVs.
1.2Minimizing Data File Size
One of the most common problems associated with running long term experiments is file size. Section 1.1.3 describes how to split multiple CVs into different experimental specifications across several different archives, which can help distribute the data and keep file sizes smaller. Another technique can be to adjust the threshold on the Advanced tab of the CV Parameters (see Figure 12).
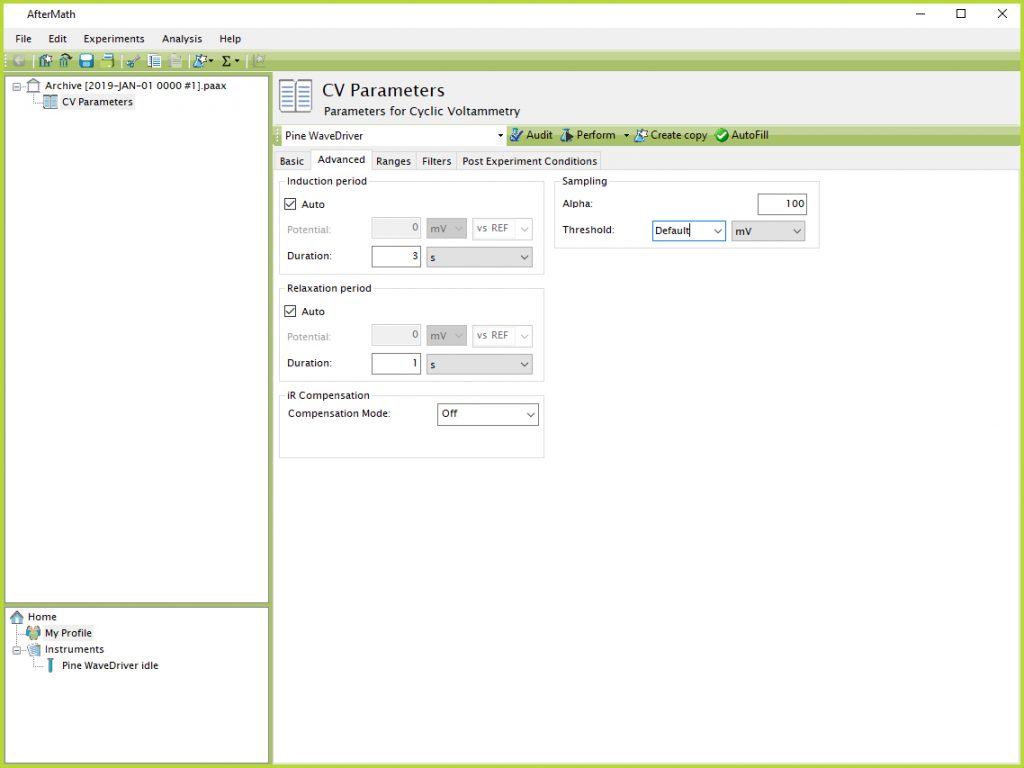
Figure 12. CV Parameters Advanced Tab Sampling Threshold Default
The "Default" setting results in a point being recorded as often as possible, which will vary depending on the scan rate but is usually around 1 mV or less. To reduce the file size, particularly during CVs in the middle of a long term experiment that are not intended to be analyzed afterwards, this threshold value can be manually increased to a larger value. This setting tells AfterMath to record data points less often when a larger potential is entered (e.g., only one point every 250 mV - see Figure 13).
NOTE: Setting a larger threshold causes AfterMath to record and save fewer data points. It does not mean AfterMath is not still performing the experiment between recorded and saved data points. The software still conducts a sweep across the entire range of parameters chosen on the Basic tab, but the saved file will consist of fewer data points based on the chosen threshold.
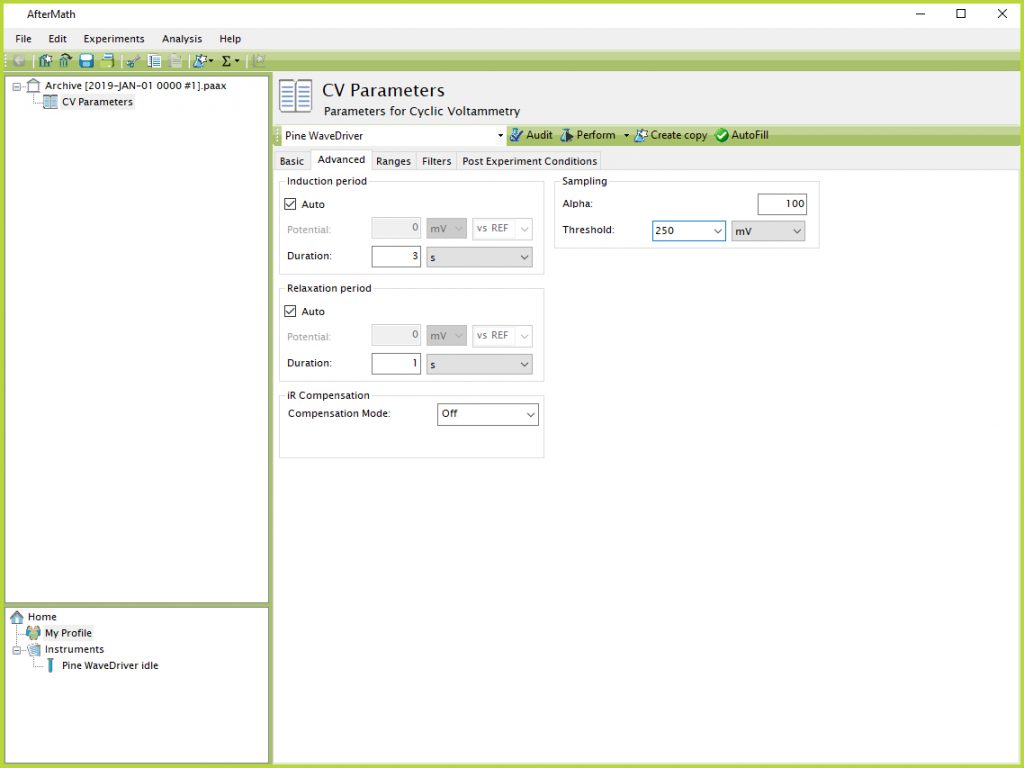
Figure 13. CV Parameters Advanced Tab Sampling Threshold 250 mV
















